Basin Attribute Extraction#
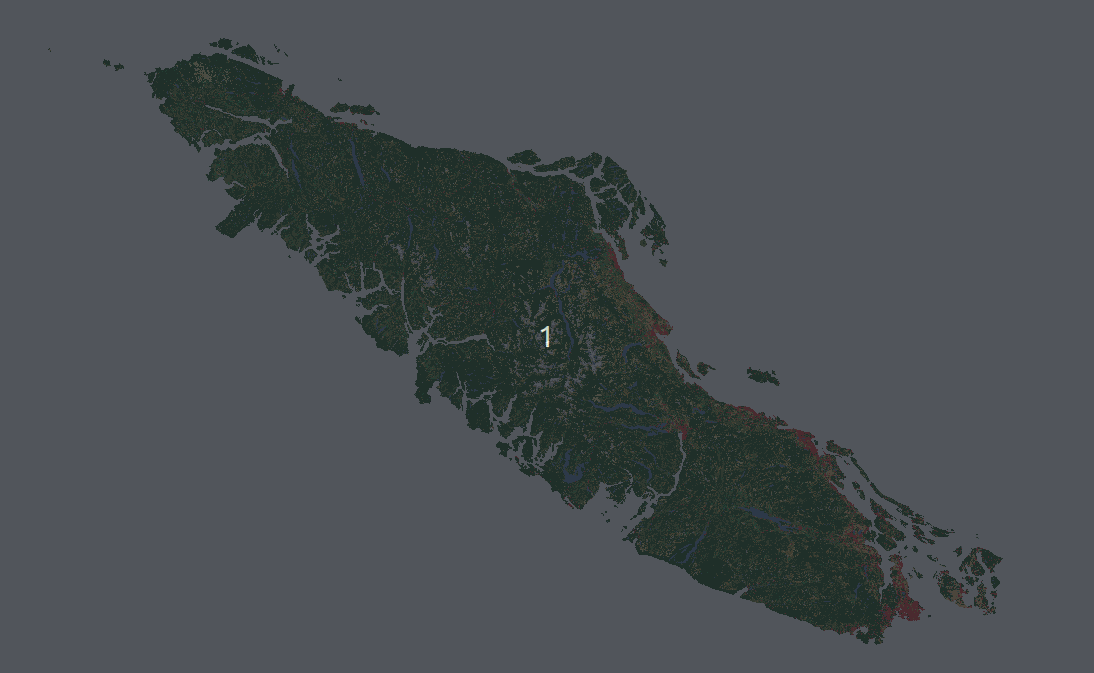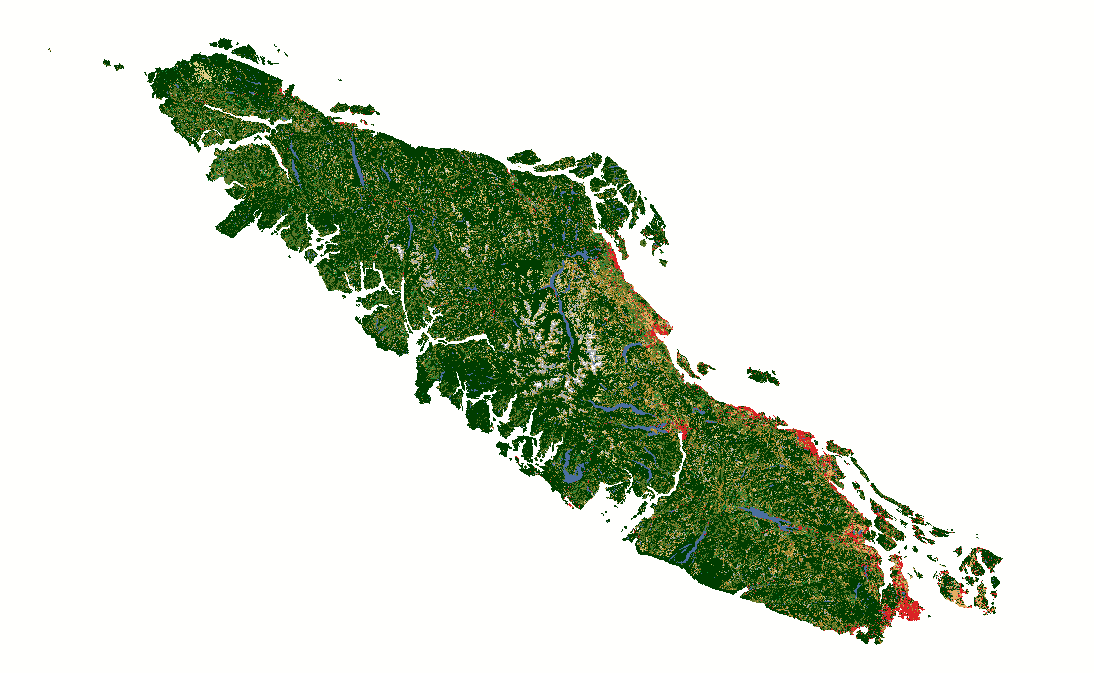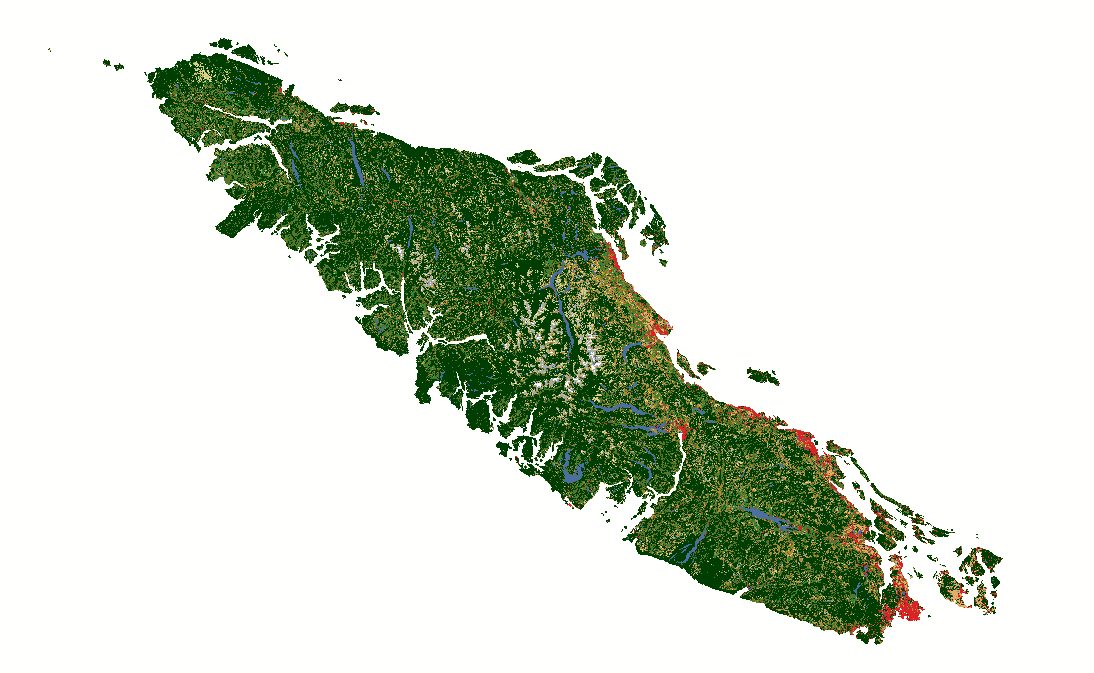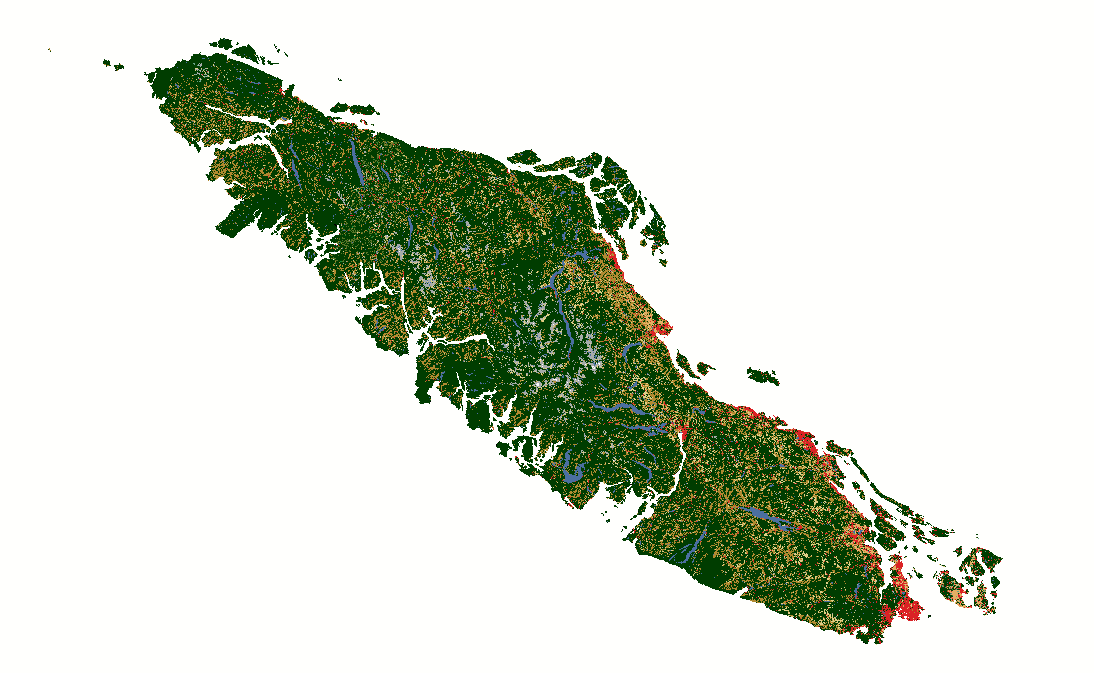
Fig. 12 North American Land Change Monitoring System latifovic2010north rasters covering Vancouver Island for 2010, 2015, and 2020.#
The final step is to capture geospatial information describing the soil, land cover, and terrain of each basin using the polygons we developed in the previous notebook. First we need to get the data and trim it to the Vancouver Island polygon.
import os
import time
import multiprocessing as mp
from utilities import retrieve_raster, clip_raster_to_basin, clip_and_reproject_NALCMS
import geopandas as gpd
import numpy as np
import pandas as pd
from shapely.geometry import box
import xarray as xr
xr.set_options(keep_attrs=True)
import rasterio as rio
import rioxarray as rxr
import metpy
from numba import jit
from scipy.stats.mstats import gmean
Land Cover#
Download and Clip NALCMS Data#
North American Coverage of the NALCMS can be downloaded from North American Land Change Monitoring System (NALCMS).
Download the files you want to work with from the link above, and use the steps below to crop the dataset to the region of interest (Vancouver Island).
Note there are NALCMS datasets for 2010, 2015, and 2020 and we’ll crop each of these in order to look at land cover changes in the next notebook.
Import the region polygon
# set the path to the polygon mask for clipping the NALCMS raster
region = 'Vancouver_Island'
mask_path = os.path.join(os.getcwd(), f'data/region_polygons/{region}.geojson')
region_polygon = gpd.read_file(mask_path)
# create a folder for the output geospatial layers
output_folder = os.path.join(os.getcwd(), 'data/geospatial_layers')
if not os.path.exists(output_folder):
os.mkdir(output_folder)
base_dir = os.path.dirname(os.getcwd())
# set the path to the downloaded NALCMS file(s)
nalcms_folder = '/media/danbot/Samsung_T51/large_sample_hydrology/common_data/NALCMS_NA/'
nalcms_files = {
2010: 'NA_NALCMS_2010_v2_land_cover_30m.tif',
2015: 'NA_NALCMS_2015_v2_land_cover_30m.tif',
2020: 'NA_NALCMS_landcover_2020_30m.tif',
}
for y in nalcms_files.keys():
output_file = f'NALCMS_{y}_clipped_3005.tif'
output_folder = os.path.join(base_dir, 'notebooks/data/geospatial_layers/')
# clip and reproject the raster and create a new file
if not os.path.exists(os.path.join(output_folder, output_file)):
clip_and_reproject_NALCMS(
nalcms_folder, output_folder,
nalcms_files[y],
mask_path, output_file, y
)
Set the DEM path which was created in notebook 2. Use the original DEM, not the (pit/depression) filled DEM.
# this should be the same path as Notebook 2
dem_dir = os.path.join(base_dir, 'notebooks/data/processed_dem/')
dem_fpath = os.path.join(dem_dir, f'{region}_3005.tif')
region_dem, dem_crs, dem_affine = retrieve_raster(dem_fpath)
Soil Permeability and Porosity#
Download and Clip GLHYMPS Data#
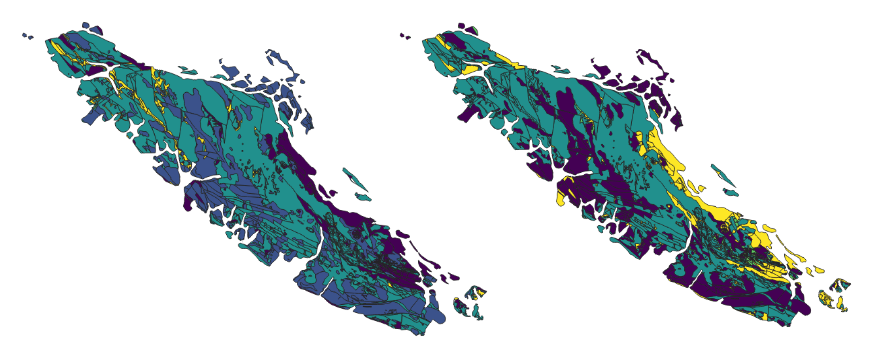
Fig. 13 The GLobal HYdrogeology MaPS (GLHYMPS) SP2_TTJNIU_2018 is global coverage of permeability and porosity in vector format.#
Download the file from here, and use the steps below to clip the data to the region of interest.
Note that this file is large, and it’s necessary to use the mask feature when opening the file using geopandas. Expect the opening and masking to take several minutes.
# glhymps is in EPSG 4326, ensure the polygon is the same CRS
region_polygon = region_polygon.to_crs(4326)
glhymps_path = 'data/geospatial_layers/GLHYMPS_Vancouver_Island.gpkg'
gldf = gpd.read_file(glhymps_path, mask=region_polygon)
Direct Attribute Extraction#
Here we load the basin polygon batch files produced in the previous notebook, and iterate through polygons to extract attributes as we go. This method is not the most performant, but the details of the process are hopefully clear.
basins_folder = os.path.join(os.getcwd(), 'data/basins/')
file = 'Vancouver_Island_basins.parquet'
# or use the geopackage
# file = 'Vancouver_Island_basins.gpkg'
# basins_df = gpd.read_file(os.path.join(basins_folder, file))
basins_df = gpd.read_parquet(os.path.join(basins_folder, file))
# basins_df = basins_df.rename({'ppt_x': 'ppt_lon_m_3005', 'ppt_y': 'ppt_lat_m_3005'}, axis=1)
print(len(basins_df))
basins_df.head()
20761
| acc | ix | jx | ppt_x | ppt_y | centroid_x | centroid_y | geometry | |
|---|---|---|---|---|---|---|---|---|
| 0 | 7072.0 | 10211 | 12194 | 1040609 | 437027 | 1039849 | 438696 | POLYGON ((1038776.917 439618.564, 1038841.431 ... |
| 1 | 234099.0 | 1603 | 6068 | 904334 | 628514 | 911107 | 617904 | POLYGON ((899882.690 629237.891, 900097.483 62... |
| 2 | 13316.0 | 3178 | 10284 | 998120 | 593478 | 999595 | 593396 | POLYGON ((998118.436 594779.872, 998458.510 59... |
| 3 | 2055.0 | 4955 | 6318 | 909895 | 553948 | 909243 | 554412 | POLYGON ((909270.083 554912.060, 909371.350 55... |
| 4 | 43332.0 | 3367 | 10446 | 1001724 | 589274 | 999161 | 589680 | POLYGON ((996824.700 592399.347, 996883.984 59... |
Table: Set of metadata and catchment attributes in the BCUB database derived from USGS 3DEP (DEM), NALCMS (land cover), and GLHYMPS (soil) datasets.
Group |
Description (BCUB label) |
Aggregation |
Units |
|---|---|---|---|
Metadata |
Pour point (geom) |
- |
decimal deg.\(^1\) |
Basin centroid point (centroid) |
- |
decimal deg. |
|
Land Cover Flag (lulc_check) |
- |
binary (0/1) |
|
———— |
—————————— |
—————– |
—————– |
Drainage Area (drainage_area_km2) |
at pour point |
\(km^2\) |
|
Elevation (elevation_m) |
spatial mean |
\(m\) above sea level |
|
Terrain |
Terrain Slope (slope_deg) |
spatial mean |
\(^\circ\) (degrees) |
Terrain Aspect (aspect_deg) |
circular mean\(^2\) |
\(^\circ\) (degrees) |
|
———— |
—————————— |
—————– |
—————– |
Cropland (land_use_crops_frac_ |
- |
||
Forest (land_use_forest_frac_ |
- |
||
Grassland (land_grass_forest_frac_ |
- |
||
Land Cover\(^3\) |
Shrubs (land_use_shrubs_frac_ |
spatial mean |
\(\%\) cover |
Snow & Ice (land_use_snow_ice_frac_ |
- |
||
Urban (land_use_urban_frac_ |
- |
||
Water (land_use_water_frac_ |
- |
||
Wetland (land_use_wetland_frac_ |
- |
||
———— |
—————————— |
—————– |
—————– |
Soil |
Permeability (permeability_logk_m2) |
geometric mean |
\(m^2\) |
Porosity (porosity_frac) |
spatial mean |
\(\%\) cover |
|
———— |
—————————— |
—————– |
—————– |
Precipitation (mean_precip_mm) |
sum |
\(mm\) |
|
Shortwave Radiation (solar_rad_Wm2) |
spatial mean |
\(W/m^2\) |
|
Climate |
Water Vapour Pressure (water_vap_Pa) |
spatial mean |
\(Pa\) |
Snow Water Equivalent (swe_kgm2) |
spatial mean |
\(kg/m^2\) |
|
Min Temperature (min_temp_C) |
spatial mean |
\(C\) |
|
Max Temperature (max_temp_C) |
spatial mean |
\(C\) |
Notes:
Geometries are formatted in the WSG84 coordinate reference system.
Spatial aspect is expressed in degrees counter-clockwise from the east direction.
The
suffix specifies the land cover dataset (2010, 2015, or 2020). Specifications on DAYMET data can be found here.
All climate parameters are mean annual values over 1980-2022.
Land Cover Data#
Land use land cover (LULC) classes are grouped as in Arsenault (2020).
def check_lulc_sum(data):
"""
Check if the sum of pct. land cover sums to 1.
Return value is 1 - sum to correspond with
a more intuitive boolean flag,
i.e. data quality flags are 1 if the flag is raised,
0 of no flag.
"""
checksum = sum(list(data.values()))
lulc_check = 1-checksum
if abs(lulc_check) >= 0.05:
print(f' ...checksum failed: {checksum:.3f}')
return lulc_check
def recategorize_lulc(data, year):
forest = (f'Land_Use_Forest_frac_{year}', [1, 2, 3, 4, 5, 6])
shrub = (f'Land_Use_Shrubs_frac_{year}', [7, 8, 11])
grass = (f'Land_Use_Grass_frac_{year}', [9, 10, 12, 13, 16])
wetland = (f'Land_Use_Wetland_frac_{year}', [14])
crop = (f'Land_Use_Crops_frac_{year}', [15])
urban = (f'Land_Use_Urban_frac_{year}', [17])
water = (f'Land_Use_Water_frac_{year}', [18])
snow_ice = (f'Land_Use_Snow_Ice_frac_{year}', [19])
lulc_dict = {}
for label, p in [forest, shrub, grass, wetland, crop, urban, water, snow_ice]:
prop_vals = round(sum([data[e] if e in data.keys() else 0.0 for e in p]), 2)
lulc_dict[label] = prop_vals
return lulc_dict
def get_value_proportions(data, year):
# create a dictionary of land cover values by coverage proportion
# assuming raster pixels are equally sized, we can keep the
# raster in geographic coordinates and just count pixel ratios
all_vals = data.data.flatten()
vals = all_vals[~np.isnan(all_vals)]
n_pts = len(vals)
unique, counts = np.unique(vals, return_counts=True)
prop_dict = {k: 1.0*v/n_pts for k, v in zip(unique, counts)}
# 15 represents cropland
# if 15 in prop_dict.keys():
# if prop_dict[15] > 0.01:
# print(f'Land cover category 15 is found: {prop_dict[15]}%')
# print(prop_dict)
prop_dict = recategorize_lulc(prop_dict, year)
return prop_dict
def process_lulc(i, basin_geom, nalcms_raster, year):
# polygon = basin_polygon.to_crs(nalcms_crs)
# assert polygon.crs == nalcms.rio.crs
if nalcms_raster.rio.crs != basin_geom.crs:
basin_geom = basin_geom.to_crs(nalcms_raster.rio.crs.to_wkt())
raster_loaded, lu_raster_clipped = clip_raster_to_basin(basin_geom, nalcms_raster)
# checksum verifies proportions sum to 1
prop_dict = get_value_proportions(lu_raster_clipped, year)
lulc_check = check_lulc_sum(prop_dict)
prop_dict[f'lulc_check_{year}'] = lulc_check
return pd.DataFrame(prop_dict, index=[i])
def check_and_repair_geometries(in_feature):
# avoid changing original geodf
in_feature = in_feature.copy(deep=True)
# drop any missing geometries
in_feature = in_feature[~(in_feature.is_empty)]
# Repair broken geometries
for index, row in in_feature.iterrows(): # Looping over all polygons
if row['geometry'].is_valid:
next
else:
fix = make_valid(row['geometry'])
try:
in_feature.loc[[index],'geometry'] = fix # issue with Poly > Multipolygon
except ValueError:
in_feature.loc[[index],'geometry'] = in_feature.loc[[index], 'geometry'].buffer(0)
return in_feature
def process_basin_elevation(clipped_raster):
# evaluate masked raster data
values = clipped_raster.data.flatten()
mean_val = np.nanmean(values)
median_val = np.nanmedian(values)
min_val = np.nanmin(values)
max_val = np.nanmax(values)
return mean_val, median_val, min_val, max_val
def get_soil_properties(merged, col):
# dissolve polygons by unique parameter values
geometries = check_and_repair_geometries(merged)
df = geometries[[col, 'geometry']].copy().dissolve(by=col, aggfunc='first')
df[col] = df.index.values
# re-sum all shape areas
df['Shape_Area'] = df.geometry.area
# calculuate area fractions of each unique parameter value
df['area_frac'] = df['Shape_Area'] / df['Shape_Area'].sum()
# check that the total area fraction = 1
total = round(df['area_frac'].sum(), 1)
sum_check = total == 1.0
if not sum_check:
print(f' Area proportions do not sum to 1: {total:.2f}')
if np.isnan(total):
return np.nan
elif total < 0.9:
return np.nan
# area_weighted_vals = df['area_frac'] * df[col]
if 'Permeability' in col:
# calculate geometric mean
# here we change the sign (all permeability values are negative)
# and add it back at the end by multiplying by -1
# otherwise the function tries to take the log of negative values
return gmean(np.abs(df[col]), weights=df['area_frac']) * -1
else:
# calculate area-weighted arithmetic mean
return (df['area_frac'] * df[col]).sum()
def process_glhymps(basin_geom, fpath):
# import soil layer with polygon mask (both in 4326)
basin_geom = basin_geom.to_crs(4326)
# returns INTERSECTION
gdf = gpd.read_file(fpath, mask=basin_geom)
# now clip precisely to the basin polygon bounds
merged = gpd.clip(gdf, mask=basin_geom)
# now reproject to minimize spatial distortion
merged = merged.to_crs(3005)
return merged
@jit(nopython=True)
def process_slope_and_aspect(E, el_px, resolution, shape):
# resolution = E.rio.resolution()
# shape = E.rio.shape
# note, distances are not meaningful in EPSG 4326
# note, we can either do a costly reprojection of the dem
# or just use the approximate resolution of 90x90m
# dx, dy = 90, 90# resolution
dx, dy = resolution
# print(resolution)
# print(asdfd)
# dx, dy = 90, 90
S, A = np.empty_like(E), np.empty_like(E)
S[:] = np.nan # track slope (in degrees)
A[:] = np.nan # track aspect (in degrees)
# tot_p, tot_q = 0, 0
for i, j in el_px:
if (i == 0) | (j == 0) | (i == shape[0]) | (j == shape[1]):
continue
E_w = E[i-1:i+2, j-1:j+2]
if E_w.shape != (3,3):
continue
a = E_w[0,0]
b = E_w[1,0]
c = E_w[2,0]
d = E_w[0,1]
f = E_w[2,1]
g = E_w[0,2]
h = E_w[1,2]
# skip i and j because they're already used
k = E_w[2,2]
all_vals = np.array([a, b, c, d, f, g, h, k])
val_check = np.isfinite(all_vals)
if np.all(val_check):
p = ((c + 2*f + k) - (a + 2*d + g)) / (8 * abs(dx))
q = ((c + 2*b + a) - (k + 2*h + g)) / (8 * abs(dy))
cell_slope = np.sqrt(p*p + q*q)
S[i, j] = (180 / np.pi) * np.arctan(cell_slope)
A[i, j] = (180.0 / np.pi) * np.arctan2(q, p)
return S, A
def calculate_circular_mean_aspect(a):
"""
From RavenPy:
https://github.com/CSHS-CWRA/RavenPy/blob/1b167749cdf5984545f8f79ef7d31246418a3b54/ravenpy/utilities/analysis.py#L118
"""
angles = a[~np.isnan(a)]
n = len(angles)
sine_mean = np.divide(np.sum(np.sin(np.radians(angles))), n)
cosine_mean = np.divide(np.sum(np.cos(np.radians(angles))), n)
vector_mean = np.arctan2(sine_mean, cosine_mean)
degrees = np.degrees(vector_mean)
if degrees < 0:
return degrees + 360
else:
return degrees
def calculate_slope_and_aspect(raster):
"""Calculate mean basin slope and aspect
according to Hill (1981).
Args:
clipped_raster (array): dem raster
Returns:
slope, aspect: scalar mean values
"""
resolution = raster.rio.resolution()
raster_shape = raster[0].shape
el_px = np.argwhere(np.isfinite(raster.data[0]))
S, A = process_slope_and_aspect(raster.data[0], el_px, resolution, raster_shape)
mean_slope_deg = np.nanmean(S)
# should be within a hundredth of a degree or so.
# print(f'my slope: {mean_slope_deg:.4f}, rdem: {np.nanmean(slope):.4f}')
mean_aspect_deg = calculate_circular_mean_aspect(A)
return mean_slope_deg, mean_aspect_deg
def get_nalcms_data(year):
nalcms_fpath = f'data/geospatial_layers/NALCMS_{year}_clipped_3005.tif'
nalcms, nalcms_crs, nalcms_affine = retrieve_raster(nalcms_fpath)
if not nalcms_crs:
nalcms_crs = nalcms.rio.crs.to_wkt()
return nalcms, nalcms_crs, nalcms_affine
Set up paths to retrieve Daymet raster files (processed in Notebook 5)#
daymet_paths, daymet_data = {}, {}
daymet_params = ['prcp', 'tmin', 'tmax', 'vp', 'swe', 'srad']
for climate_param in daymet_params:
fpath = f'data/daymet_data/{climate_param}_mosaic_3005.tiff'
daymet_raster, daymet_crs, daymet_affine = retrieve_raster(fpath)
daymet_data[climate_param] = daymet_raster
# to test, try on a few samples, note how basins_df is sliced in the for .. statement next
# to run the whole batch, remove the [:n_samples] slice
# n_samples = 11
# results_fname = file.replace('_basins.gpkg', '_attributes.gpkg')
results_fname = file.replace('_basins.parquet', '_attributes.parquet')
results_path = os.path.join('data', results_fname)
# filter the basins_df dataframe to exclude basins already processed
if os.path.exists(results_path):
# if 'ppt_y' in basins_df.columns:
# basins_df = basins_df.rename({'ppt_y': 'ppt_lat_m_3005'}, axis=1)
# if 'ppt_x' in basins_df.columns:
# basins_df = basins_df.rename({'ppt_x': 'ppt_lon_m_3005'}, axis=1)
if results_path.endswith('.parquet'):
existing_results = gpd.read_parquet(results_path)
# existing_results[['ppt_lon_m_3005', 'ppt_lat_m_3005']] = existing_results[['ppt_lon_m_3005', 'ppt_lat_m_3005']].round(0).astype(int)
else:
existing_results = gpd.read_file(results_path)
# print(asdf)
n_basins, n_results = len(basins_df), len(existing_results)
basins_df = basins_df[~basins_df.set_index(match_cols).index.isin(existing_results.set_index(match_cols).index)]
print(f'{n_results} basins already processed. {len(basins_df)} remaining.')
def update_results(new_data):
new_df = gpd.GeoDataFrame(new_data, crs=3005)
if not os.path.exists(results_path):
print(f'Processed {len(new_df)} basins ({results_fname}).')
if results_path.endswith('.parquet'):
new_df.to_parquet(results_path, index=False)
else:
new_df.to_file(results_path, index=False)
else:
if results_path.endswith('.parquet'):
existing_data = gpd.read_parquet(results_path)
output = gpd.GeoDataFrame(pd.concat([existing_data, new_df]))
output.to_parquet(results_path, index=False)
else:
existing_data = gpd.read_file(results_path)
output = gpd.GeoDataFrame(pd.concat([existing_data, new_df]))
output.to_file(results_path, index=False)
print(f'Processed {len(new_df)} basins ({results_fname}) ({len(output)} processed).')
all_basin_data = []
t0 = time.time()
last_index = basins_df
for i, row in basins_df.iterrows():
basin_data = {}
basin_data['region'] = region
basin_data['id'] = i
basin_polygon = basins_df[basins_df.index == i].copy()
clip_ok, clipped_dem = clip_raster_to_basin(basin_polygon, region_dem)
for y in [2010, 2015, 2020]:
nalcms, nalcms_crs, nalcms_affine = get_nalcms_data(y)
land_cover = process_lulc(i, basin_polygon, nalcms, y)
land_cover = land_cover.to_dict('records')[0]
# print(' lulc complate')
basin_data.update(land_cover)
soil = process_glhymps(basin_polygon, glhymps_path)
porosity = get_soil_properties(soil, 'Porosity')
permeability = get_soil_properties(soil, 'Permeability_no_permafrost')
basin_data['Permeability_logk_m2'] = round(permeability, 2)
basin_data['Porosity_frac'] = round(porosity, 5)
slope, aspect = calculate_slope_and_aspect(clipped_dem)
# print(f'aspect, slope: {aspect:.1f} {slope:.2f} ')
basin_data['Slope_deg'] = slope
basin_data['Aspect_deg'] = aspect
mean_el, median_el, min_el, max_el = process_basin_elevation(clipped_dem)
basin_data['median_el'] = median_el
basin_data['mean_el'] = mean_el
basin_data['max_el'] = max_el
basin_data['min_el'] = min_el
# geojson only supports one geometry column
basin_data['geometry'] = row.geometry
basin_data['ppt_x'] = row['ppt_x']
basin_data['ppt_y'] = row['ppt_y']
basin_data['centroid_x'] = row['centroid_x']
basin_data['centroid_y'] = row['centroid_y']
for param in daymet_params:
clipped_data = daymet_data[param].rio.clip(basin_polygon.geometry, all_touched=True)
spatial_mean = round(clipped_data.mean(dim=['y', 'x']).item(), 1)
basin_data[param] = spatial_mean
all_basin_data.append(basin_data)
# don't update at the very start
# only update periodically
# update at the end!
if ((i > 0) & (i % 100 == 0)) | (i >= max(basins_df.index.values)):
t_int = time.time()
ii = i
if ii == 0:
ii = 1
ut = (t_int - t0) / ii
print(f' ...processed {i}/{len(basins_df)} ({ut:.2f} s/basin). Updating results file.')
update_results(all_basin_data)
all_basin_data = []
...processed 100/20761 (1.04 s/basin). Updating results file.
Processed 101 basins (Vancouver_Island_attributes.parquet).
...processed 200/20761 (0.99 s/basin). Updating results file.
Processed 100 basins (Vancouver_Island_attributes.parquet) (201 processed).
...processed 300/20761 (1.00 s/basin). Updating results file.
Processed 100 basins (Vancouver_Island_attributes.parquet) (301 processed).
...processed 400/20761 (1.01 s/basin). Updating results file.
Processed 100 basins (Vancouver_Island_attributes.parquet) (401 processed).
...processed 500/20761 (1.02 s/basin). Updating results file.
Processed 100 basins (Vancouver_Island_attributes.parquet) (501 processed).
...processed 600/20761 (1.03 s/basin). Updating results file.
Processed 100 basins (Vancouver_Island_attributes.parquet) (601 processed).
...processed 700/20761 (1.04 s/basin). Updating results file.
Processed 100 basins (Vancouver_Island_attributes.parquet) (701 processed).
...processed 800/20761 (1.05 s/basin). Updating results file.
Processed 100 basins (Vancouver_Island_attributes.parquet) (801 processed).
...processed 900/20761 (1.06 s/basin). Updating results file.
Processed 100 basins (Vancouver_Island_attributes.parquet) (901 processed).
...processed 1000/20761 (1.07 s/basin). Updating results file.
Processed 100 basins (Vancouver_Island_attributes.parquet) (1001 processed).
...processed 1100/20761 (1.07 s/basin). Updating results file.
Processed 100 basins (Vancouver_Island_attributes.parquet) (1101 processed).
Processing time is roughly 0.8s/basin on a ~2018 Intel i7-8850H CPU @ 2.60GHz. Processing time is proporti onal to the size of clipped DEM, and if you can use JIT in the process_slope_and_aspect function, it yields roughly 3X performance improvement.
